Plotting Fit Results#
import ROOT
import os
from pyamptools.utility.plotgen_utils import draw_histograms
from pyamptools import atiSetup
from IPython.display import Image
PYAMPTOOLS_HOME = os.environ["PYAMPTOOLS_HOME"]
atiSetup.setup(globals(), use_fsroot=True)
ROOT.ROOT.EnableImplicitMT() # REMOVE THIS WHEN DEBUGGING
Welcome to JupyROOT 6.28/06
atiSetup| jupyter-book called python3.9
------------------------------------------------
atiSetup| MPI is disabled
atiSetup| GPU is disabled
------------------------------------------------
atiSetup| Loading library libIUAmpTools.so ............ ON
atiSetup| Loading library libAmpTools.so .............. ON
atiSetup| Loading library libAmpPlotter.so ............ ON
atiSetup| Loading library libAmpsDataIO.so ............
ON
atiSetup| Loading library libFSRoot.so ................ ON
atiSetup| Loading library libAmpsGen.so ............... OFF
------------------------------------------------
------------------------------------------------
atiSetup| Saved aliases found in /d/grid17/ln16/PyAmpTools/src/pyamptools/.aliases.txt, attempting to load...
atiSetup| minor warning: Unable to alias omegapiAngles - doesn't exist under ROOT namespace
atiSetup| minor warning: Unable to alias URConfig - doesn't exist under ROOT namespace
atiSetup| minor warning: Unable to alias URtypes - doesn't exist under ROOT namespace
atiSetup| minor warning: Unable to alias FSFitFunctions - doesn't exist under ROOT namespace
atiSetup| minor warning: Unable to alias FSFitPrivate - doesn't exist under ROOT namespace
After performing a MLE fit a FitResults object is created with a .fit extension. This file can be read in using the AmpTools PlotGenerator class. In this class, Amplitudes can be turned on/off and amplitude weights can be extracted for a particular amplitude combination. These weights can then be used for plotting purposes.
This tutorial will go over how we can use FSRoot and RDataFrames to produce fit results.
Numpy is used as an intermediary between PlotGenerator and RDataFrame. This choice is made so that users can makes plots in the Python ecosystem if they wish instead of going through ROOT. It is important to remark that RDataFrame does not perform any copying and reads directly from the Numpy array.
Some macros can be loaded ( mirroring FSRoot macros / math ) that calculates various kinematic quantites (i.e. mass and helicity angles). We can load them as follows. We will return to their definitions in a bit.
from pyamptools.utility.rdf_macros import loadMacros
loadMacros()
Loading FSMath macros from /d/grid17/ln16/PyAmpTools/utility/RDF_userDefFuncs.cc
We can also use PyROOT to load headers files. Here we will load the gluex_style and set the plotting style
### IF USING ROOT TO PLOT - CAN SET GLUEX STYLE HERE ###
gInterpreter.ProcessLine('#include "gluex_style.h"')
gluex_style = ROOT.gluex_style() # Returns TStyle
gluex_style.SetPadRightMargin(0.08)
gluex_style.cd()
Lets set some additional alias and enable multi-threading for faster results
Lets define the location of the .fit file and the name of a .pdf file to dump drawn histograms to
fit_results = f"{PYAMPTOOLS_HOME}/tests/samples/SIMPLE_EXAMPLE/result.fit"
hist_output_name = "result" # dumps results to a pdf file with this name, extension will be appended
Lets load the .fit file into a FitResults object
results = FitResults(fit_results)
if not results.valid():
print(f"Invalid fit result in file: {fit_results}")
exit()
=================================================================
| ^ |
| / \ Version: v0.15.3-2-g0753-dirty |
| /---\ |
| / \ GDouble: 8 bytes |
| / \ MP MPI: NO |
| ------- GPU: NO |
| | |
| | doi.org/10.5281/zenodo.5039377 |
| | OOLS |
=================================================================
As always, we have to register the requested amplitudes and the datareaders. We can then create a PlotGenerator instance. The order matters as PlotGenerator uses these amplitudes and datareaders.
plotGen = PlotGenerator(results)
[ AmplitudeManager ]:
Creating AmplitudeManager for the reaction: etapi
particle index assignment: Beam -> 0
particle index assignment: Proton -> 1
particle index assignment: Eta -> 2
particle index assignment: Pi0 -> 3
We can book histograms in the dictionary format shown below. For now, we will just make 1D histograms of several kinematic quantities. We use macros inspired and using FSRoot format to compute quantities. The macros are defined in utils/RDFmacros.py, see here for source code.
For example, MASS(ETA,PI0) would compute the invariant mass of the ETA, PI0 system where ETA and PI0 are defined particles. The order of the particles matters and should match what is seen in the configuration file.
In RDataFrame, histograms are first booked and lazily evaluated, to define as much computation as possible before a request for a result is made.
############### BOOKEEPING ################
## START WITH 1D HISTS SO WE CAN REUSE FUNCTION VALUES! ##
## FOR THIS EXAMPLE, WILL NOT INCLUDE 2D PLOTS SO WE CAN STACK 1D HISTS FOR FIT RESULTS ##
HISTS_TO_BOOK = {
# 1D Hists
# HistName: [ xname, Function, title, n_bins, x-min, x-max, drawOptions]
"Metapi": ["Metapi", "MASS(ETA,PI0)", ";M(#eta#pi);Events", 50, 1.04, 1.72, ""],
"Meta": ["Meta", "MASS(ETA)", ";M(#eta);Events", 50, 0.49, 0.61, ""],
"Mpi0": ["Mpi0", "MASS(PI0)", ";M(#pi^{0});Events", 50, 0.1, 0.18, ""],
"cosGJ": ["cosGJ", "GJCOSTHETA(ETA,PI0,RECOIL)", ";cos(#theta_{GJ});Events", 50, -1, 1, ""],
"cosHel": ["cosHel", "HELCOSTHETA(ETA,PI0,RECOIL)", ";cos(#theta_{HEL});Events", 50, -1, 1, ""],
"phiHel": ["phiHel", "HELPHI(ETA,PI0,RECOIL,GLUEXBEAM)", ";#phi_{HEL};Events", 50, -1, 1, ""],
# 2D Hists
# HistName: [ xname, xfunction, title, nx_bins, x_min, x_max, yname, yfunction, ny_bins, y_min, y_max, drawOptions]
# "cosHelvsMass": [ "Metapi", "MASS(ETA,PI0)", "M(#eta#pi) vs cos(#theta_{hel})", 100, 1.04, 1.72, "cosHel", "GJCOSTHETA(ETA,PI0,GLUEXBEAM)", 100, -1, 1, "COLZ"],
}
############## SETUP ##############
particles = ["GLUEXBEAM", "RECOIL", "ETA", "PI0"]
draw_histograms is an example implemenation of how to take the dictionary of histogram specification, fill histograms, and draw the result. See draw_histograms API for more information and source code. We can pass in an amplitudes string containing a space separated string of wavesets which are themselves semicolon separated list of amplitudes and draw them all.
amplitudes = "all resAmp1 resAmp2 resAmp3 resAmp1;resAmp2"
draw_histograms(results, hist_output_name, particles, HISTS_TO_BOOK, amplitudes, output_format="png")
Loaded data into PlotGenerator...
>> Plotting waveset: all
>> Keeping all amplitudes: { "resAmp1", "resAmp2", "resAmp3" }
>> Plotting waveset: resAmp1
>> Plotting waveset: resAmp2
>> Plotting waveset: resAmp3
>> Plotting waveset: resAmp1;resAmp2
Creating AmplitudeManager for the reaction: etapi
particle index assignment: Beam -> 0
particle index assignment: Proton -> 1
particle index assignment: Eta -> 2
particle index assignment: Pi0 -> 3
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Info in <TCanvas::Print>: png file result_all.png has been created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Info in <TCanvas::Print>: png file result_resAmp1.png has been created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Info in <TCanvas::Print>: png file result_resAmp2.png has been created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Info in <TCanvas::Print>: png file result_resAmp3.png has been created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Warning in <TH1D::Sumw2>: Sum of squares of weights structure already created
Info in <TCanvas::Print>: png file result_resAmp1_resAmp2.png has been created
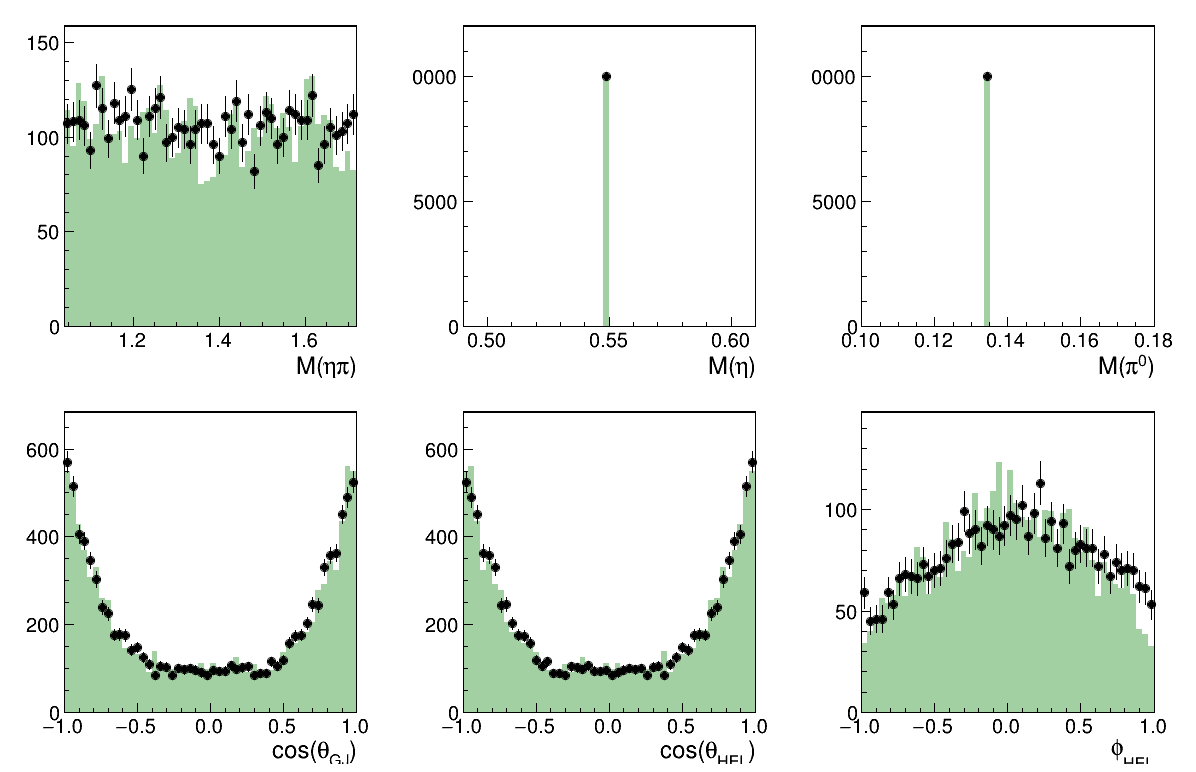
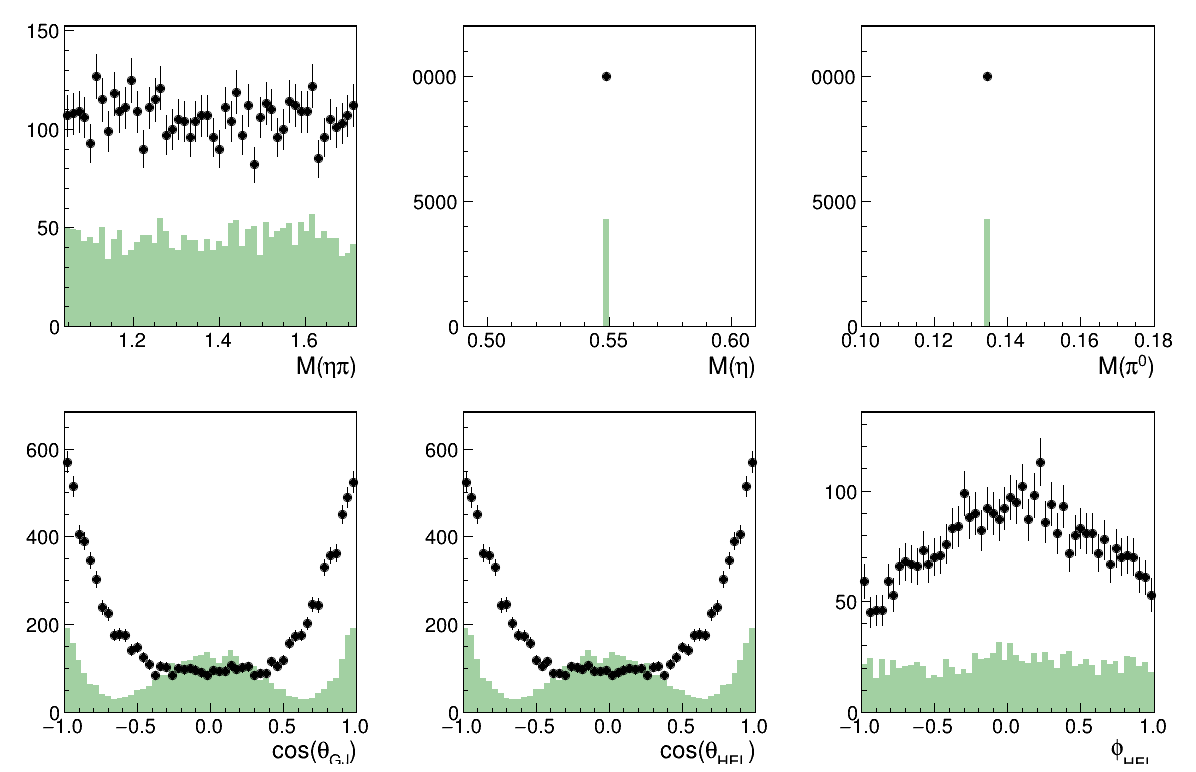
Overall Fit results
Image(filename="result_all.png")
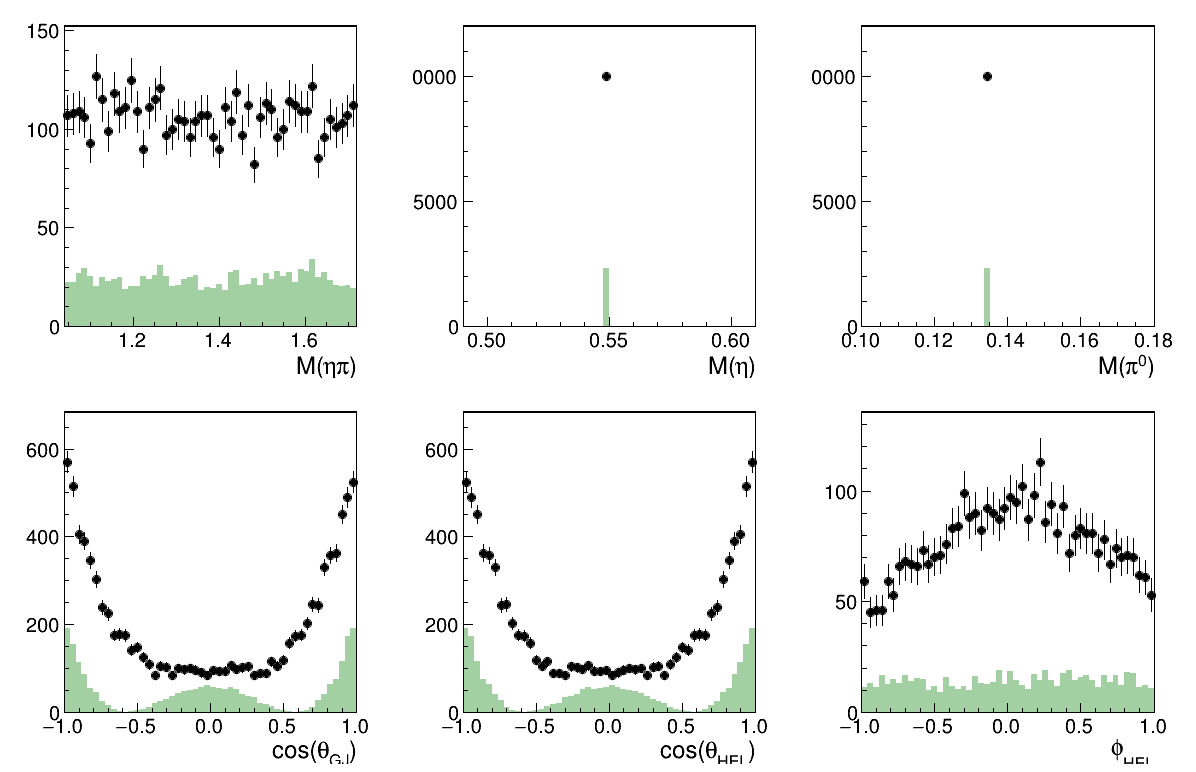
Only the L=2 M=0 wave
Image(filename="result_resAmp1.png")
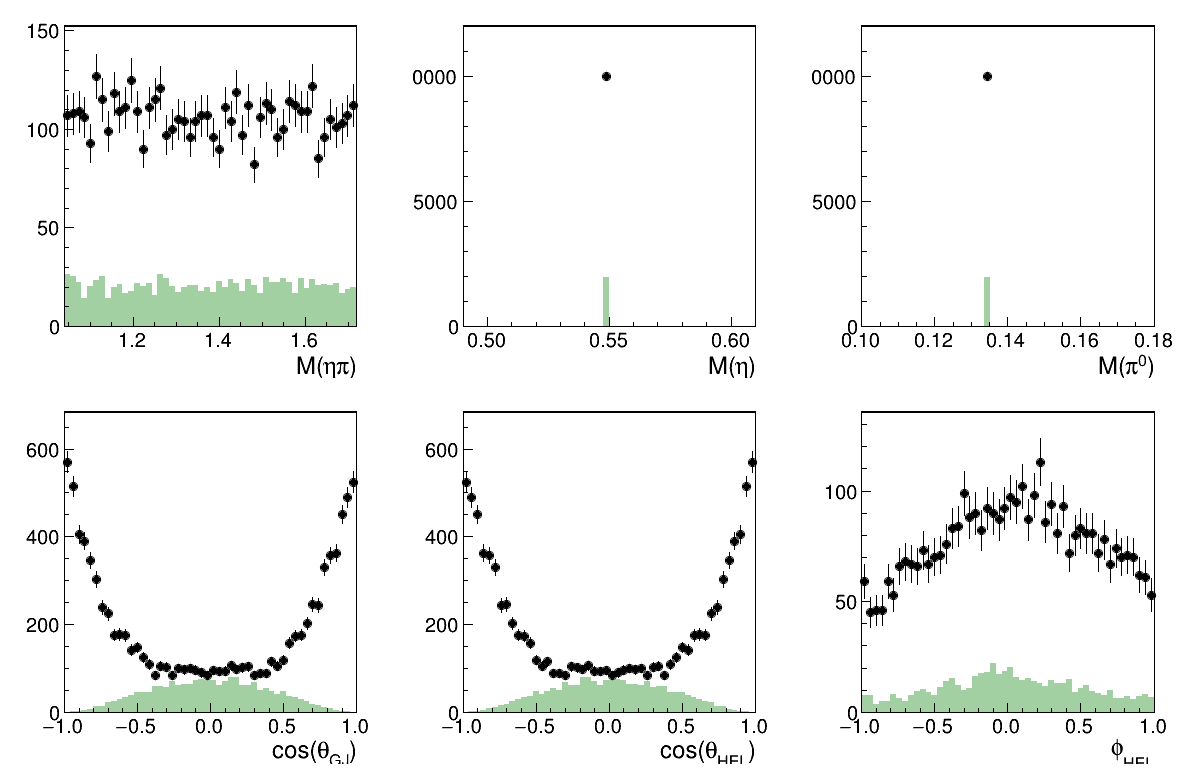
Only the L=2 M=2 wave
Image(filename="result_resAmp2.png")
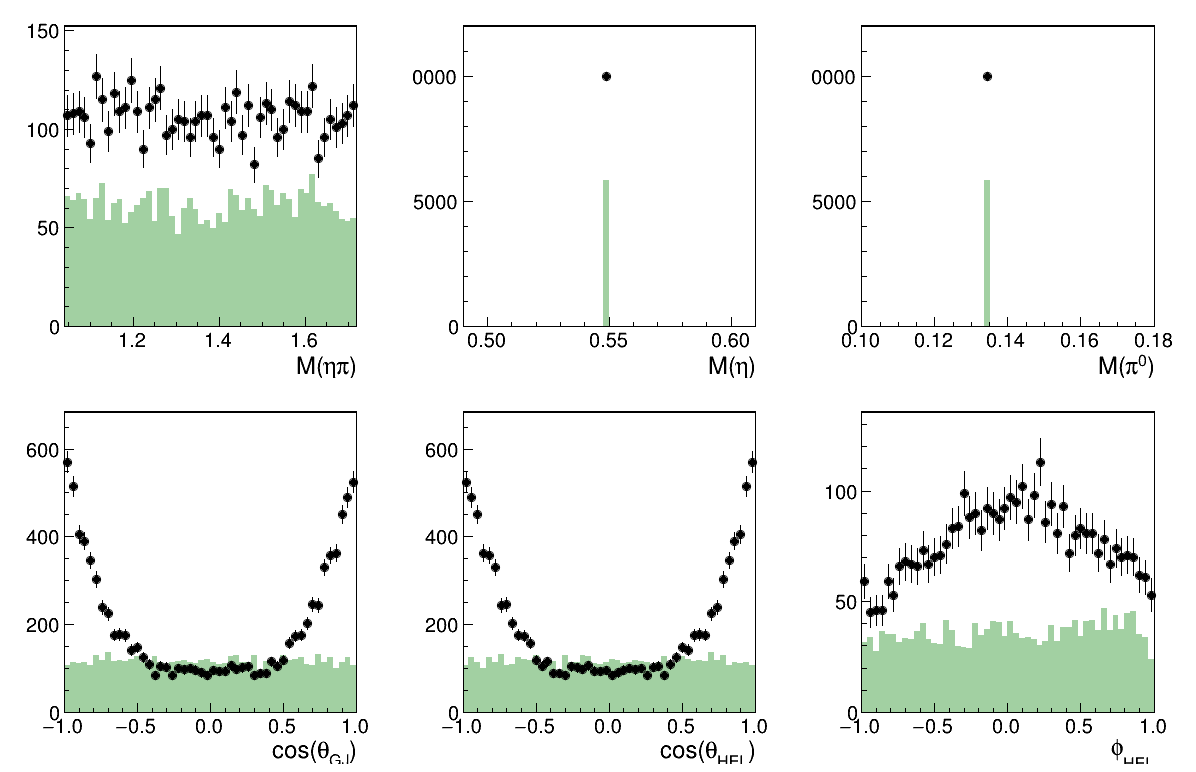
Only the L=0 M=0 wave
Image(filename="result_resAmp3.png")
Coherent sum of (L=2, M=0) (L=2, M=0) waves
Image(filename="result_resAmp1_resAmp2.png")
# cleanup again
!rm result*.png